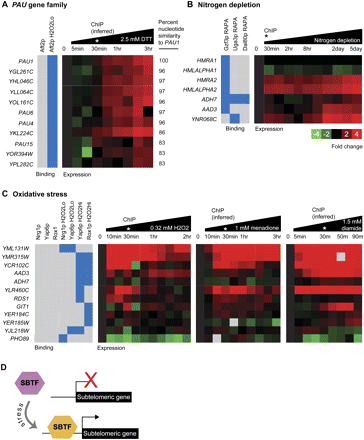
Dynamic binding and expression profiles suggest models of SBTF function and link SBTFs to poorly characterized genes at subtelomeres. TF binding profiles were matched to expression profiles gathered under similar environmental perturbations. (A) Aft2p binds upstream of 12 subtelomeric PAU genes under oxidative stress conditions (blue boxes). Heatmap shows induction of the PAU genes in oxidative stress conditions. (B) In the presence of rapamycin, which simulates nitrogen depletion by antagonizing the TOR kinases (Magasanik and Kaiser 2002), Gzf3p, Uga3p, and Dal80p bind upstream of genes that are induced under conditions of nitrogen limitation. (C) Genes targeted by Nrg1p, Yap6p, and/or Rox1p under hydrogen peroxide but not in untreated conditions are up-regulated in response to three oxidative stress agents (hydrogen peroxide, menadione, and diamide). For A–C, the asterisk (*) indicates the approximate time that the binding data were collected. (D) SBTFs that display a preference for the subtelomere only in stress conditions may be positive regulators of gene expression (see text). Expression heatmaps all use scale shown in B.











