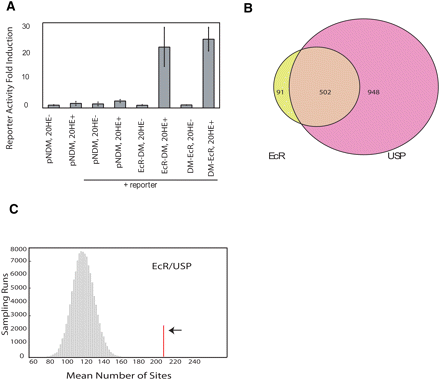
(A) ECR-DamMyc(DM) and DamMyc-ECR fusion proteins transduce the 20-HE signal in Kc cells (L57-3-11) using a wild-type copy of the endogenous EcR gene, showing that both fusion proteins are functional. pnDM (pnDamMyc) is the vector containing the N terminus Dam protein, and the lacZ reporter was used to measure beta-galactidose activity. (B) Overlap of significantly bound regions by ECR and USP at P < 0.001. (C) Enrichment of computationally predicted ECR/USP-binding sites in ECR/USP-bound regions. The red line indicates the number of computationally identified ECR/USP sites within the binding regions (P < 0.0001). The histogram shows the distribution (sampled from 100,000 runs) of putative ECR/USP sites number within the same number of randomly selected 4-kb windows.











