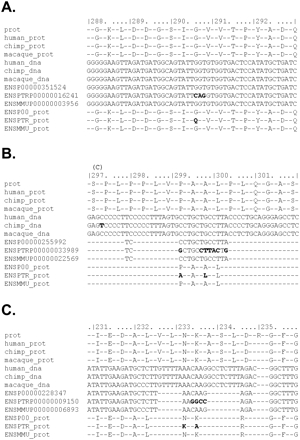
Sequence error revealed by alignments. Genome sequence errors in the template chimpanzee genome sequence used in test set 1 dominate the signal for positive selection in chimpanzee. We generated meta-alignments of each of the 49 genes that we reanalyzed for this study, which compare our alignments (with the new bioinformatics pipeline) with the alignments originally provided by the authors of the studies of test set 1 (Bakewell et al. 2007) (see Methods). Three examples from test set 1 are presented, where clusters of chimpanzee-specific divergent sites within a codon appear to be causing a false-positive signal of a chimpanzee PSG: (A) HELZ, (B) KRBA1 (NP_115923.1), and (C) POLR3B. In our realignment, these clusters of divergent sites disappear. There are 13 lines in each meta-alignment. Line 1 is the reference Ensembl human protein sequence for the gene. Lines 2–4 show the protein translations of our DNA alignments for human, chimpanzee, and macaque, respectively, and lines 5–7 show the DNA alignments themselves. Lines 8–10 show the corresponding alignment published in test set 1 (Bakewell et al. 2007). Lines 11–13 show translations of the DNA from test set 1. ENSP (lines 8,11) and ENSPTR (lines 9,12) refer to human and chimpanzee, respectively. Positions within the protein alignment that do not match the protein consensus are highlighted. Sequence differences are highlighted above the alignment. “C” indicates a synonymous chimpanzee divergent site. Macaque divergent sites are not highlighted.











