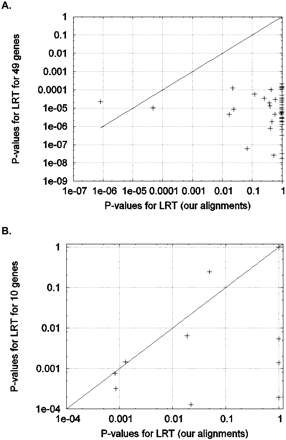
Positive selection in the chimpanzee lineage for each gene from each test set (see Methods). (A) Likelihood ratio test analysis of genes comparing our alignments with the alignments of test set 1: The majority of the 49 genes that we reanalyzed from test set 1 showed significant P-values (<0.0001) for positive selection in chimpanzee when we analyzed them using the alignments provided to us by the authors. However, our alignments indicate no positive selection in chimpanzee (P > 0.05) at all genes except for RNF130 and USP44. As described in the text, there is evidence that RNF130 is a false positive that emerges both from our analysis pipeline and that of test set 1. (B) The same analysis finds that five of the 10 signals of chimpanzee PSGs that we reanalyzed from test set 2 do not replicate after our realignment.











