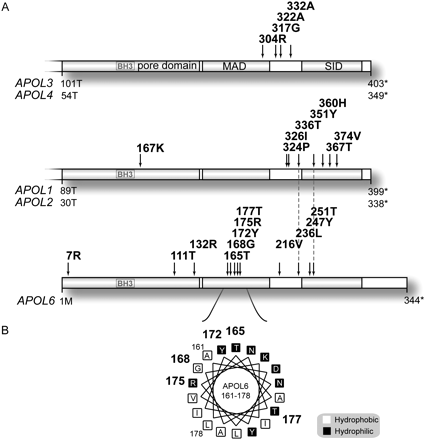
Codons under positive selection in primate APOL genes, and a cluster of rapidly evolving codons on an APOL6 MAD alpha helix. (A) We used maximum-likelihood-based tests to estimate positive selection in APOL1, APOL2, and APOL2.1 (combined); APOL3 and APOL4 (combined); and APOL6. We did not analyze APOL5 due to lack of available sequences. The genes are shown to scale and are aligned with one another. The dotted lines show two codons that are under positive selection in both APOL1/L2/L2.1 and APOL6. Gene coordinates and amino acids are shown for human genes, and the isoforms shown include APOL1 isoform a and APOL3 isoform1. APOL2.1 is not shown in the figure. Asterisks indicate stop codons. (B) A group of rapidly evolving codons in the APOL6 MAD tightly cluster on one face of an alpha helix, the predicted structure for the region. Rapidly evolving codons are shown in bold. Sites are also shaded according to hydrophobicity, and the helix has 3.6 amino acids per turn. The rapidly evolving sites of the MAD are located at the base of a pH-sensitive hinge structure. The helix appears to be amphipathic, and rapidly evolving sites appear to be at the hydrophobic/hydrophilic interface of the helix. MAD: membrane-addressing domain; SID: SRA-interacting domain.











