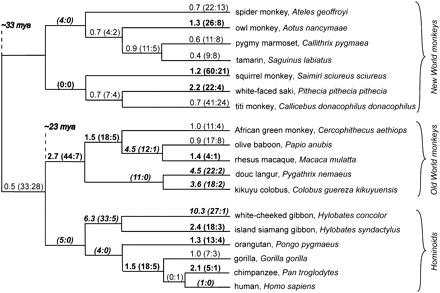
Lineage-specific evidence for positive selection in primate APOL6. A cladogram is shown with maximum-likelihood estimates of lineage-specific dN/dS during primate evolution. Numbers in parentheses are the estimated number of nonsynonymous and synonymous changes, respectively, for each branch. Values of dN/dS are also colored according to the class of dN/dS to which they are predicted to belong (by GABranch analysis): Bolded values belong to class dN/dS = 1.49; bolded, italicized values belong to class dN/dS = 5.92; and nonbolded values belong to class dN/dS = 0.63. Branch lengths in the figure are not proportional to time.











