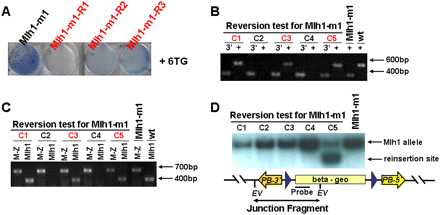
Reversibility of Mlh1-m1 mutation with PBase. (A) Low-density survival assay demonstrating 6-TG sensitivity of three revertant clones. (B) Genomic PCR analysis of 96 reversal candidates with primers for the PB3′ IR and sequences upstream of insertion site of Mlh1 (marked as “3′”) detected the transposon/host junction of the expected size. Representative candidate clones including three reverted ones are shown. The genomic PCR product fragments (marked as “+”) are absent in two mutant clones (C2 and C4) but were amplified from three revertants (C1, C3, and C5). (C) Expression of wild-type Mlh1 and fusion Mlh1-lacZ transcripts in gene-trap mutants. RT-PCR with primers for exon2 of Mlh1 and lacZ (marked as “M-Z”) detected fusion transcripts of the expected size from all 96 clones. The wild-type transcripts (marked as “Mlh1”) are absent in clones C2 and C4 but are found in the three revertants (C1, C3, and C5). (D) Southern blot analysis of Mlh1-m1 revertant clones identifies a new integration of the transposon in revertant clone 5. EV, EcoRV. Probe, LacZ.











