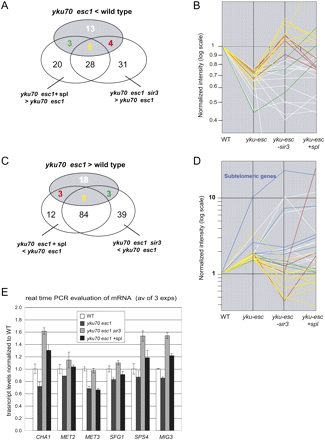
Impairing SIR complex function rescues half of the genes misregulated upon deletion of telomere anchoring pathways. (A) Fifty-three percent of the down-regulated genes in a yku70 esc1 (GA-2768) strain are rescued either by deleting sir3 (GA-2985, 43%) or by inhibiting Sir2 (39%) with splitomicin (20 μM) or both (29%). (B) mRNA levels detected on microarrays are shown in the four indicated conditions (wild type, yku70 esc1, splitomicin-treated yku70 esc1, and yku esc1 sir3 strains) for genes down-regulated >1.5-fold between wild-type and the isogenic yku70 esc1 strain. For each of these genes mRNA levels are normalized to wild-type levels. (C) Forty-four percent of the up-regulated genes in the yku70 esc1strain are rescued by deleting SIR3 (34%) or by inhibiting Sir2 (34%) with splitomicin (20 μM) or both (25%). (D) As in B, for genes up-regulated >1.5-fold between wild-type and the isogenic yku70 esc1 strain. (E) mRNA levels were determined by quantitative real-time PCR on mRNA isolated from the indicated strains which are identical to those used for the genome-wide analysis. Normalization is by geomean to NUP1 and NUP159 transcripts as in Taddei et al. (2006). Error bars correspond to SEM. Similar results were obtained for YLR156W, although unlike the others, in the presence of splitomicin expression was elevated above the wild-type levels (data not shown).











