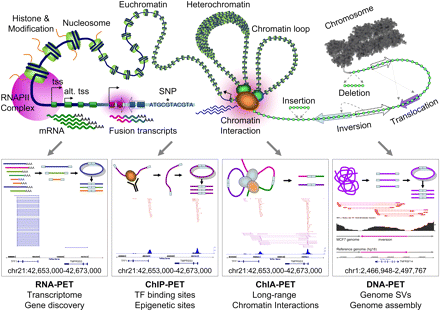
PET applications to address genome biology questions. Cells have many different mechanisms for processing, modifying, controlling, and transducing information encoded in the genome. The PET technology can be applied to investigate many questions regarding nuclear processes, such as transcriptomes by RNA-PET, transcription and epigenetic regulation by ChIP-PET and ChIA-PET, as well as genome structural variation by DNA-PET. Examples of PET data from GIS-PET (an early version of RNA-PET), ChIP-PET, and ChIA-PET experiments of human breast cancer MCF-7 cells with estrogen induction treatment at the TFF1 locus (chr21:42,653,000-42,673,000) are shown: the high level of TFF1 gene expression and the low level of TMPRSS3 gene expression; the ERα binding at the TFF1 promoter and enhancer sites; and the long-range chromatin interactions between the two ERα binding sites. An example of DNA-PET data at the TNFRSF14 locus in the genome of MCF-7 cells shows an inversion event detected by two clusters of discordant DNA-PET cluster mapping.











