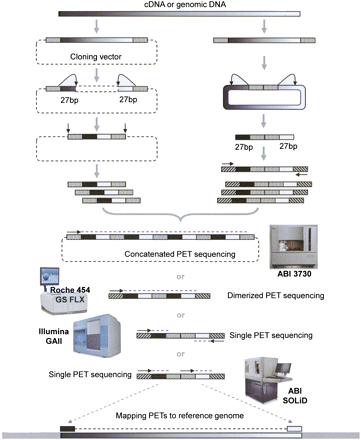
Schematic view of PET methodology. PET construction may be carried out through cloning-based or cloning-free procedures. In the cloning-based procedure, DNA fragments are ligated to cloning vector to introduce restriction sites such as EcoP15I to the 5′ and 3′ ends of insert DNA and link the two ends by a vector backbone. This is transformed into E. coli cells as a DNA library. EcoP15I digestion of the library will result in tag–vector–tag structures, which are re-ligated to form a single-PET library and further digested to release the PET constructs. In the cloning-free protocol, linker oligonucleotides containing EcoP15I sites are directly ligated to DNA fragments, followed by circularization, and then digestion by EcoP15I to release the PET constructs. The resulting PET constructs can be analyzed by concatemer sequencing using Sanger capillary instrument, dimerization sequencing using Roche 454 GS, paired-end sequencing using Illumina GA or Applied Biosystems SOLiD. The PET sequences are then mapped to reference genome sequences to demarcate the boundaries of the target DNA fragments.











