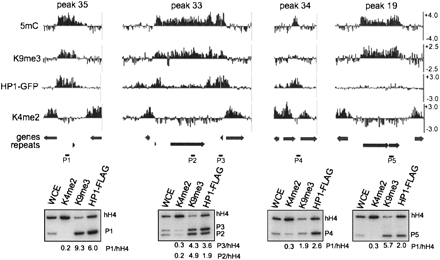
Complex localization patterns of HP1. Microarray data from immunoprecipitation experiments using antibodies to 5mC, H3K9me3, green fluorescent protein (for HP1-GFP; HP1), and H3K4me2 are shown as log2 values for 5mC peaks 35, 33, 34, and 19 (Supplemental Table 1). The position of predicted open reading frames (genes) and repeats are indicated below. Conventional ChIP was performed for each region using antibodies to H3K4me2, H3K9me3, and FLAG (for HP1-FLAG) to validate the microarray data. PCR products obtained using whole cell extracts (WCE), or the indicated immunoprecipitate fraction as the template, were resolved by gel electrophoresis. The black bars labeled P1–P5 depict the position of the expected PCR products within each region. The coding region of histone H4 was used as an internal euchromatin control. The labels to the right of each autoradiograph identify specific PCR products. The enrichment of each PCR product relative to the PCR product for H4 to is shown below.











