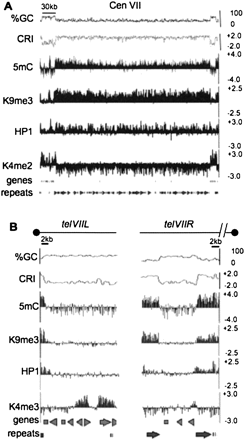
Chromatin modification profile for the LGVII centromere and telomeres. For (A) N. crassa LGVII centromere and (B) N. crassa chromosome ends, nucleotide composition is shown as the moving average of %GC and CRI calculated for 500 bp windows with 100 bp steps at the top of the plot. Enrichment values for MeDIP and ChIP-chip experiments are shown as log2 values indicated on the y-axis (right) for immunoprecipitation experiments using antibodies to 5mC, H3K9me3, green fluorescent protein for HP1-GFP (HP1) and H3K4me2. The position of predicted open reading frames (genes) and repeats are indicated below. The scale bars on the top indicate 30 kb and 2 kb for (A) and (B), respectively. The black dots above each plot in (B) indicate the position of the telomere. The broken line above telVIIR indicates that sequences containing the telomere repeats for telVIIR are missing from the current LGVII sequence assembly.











