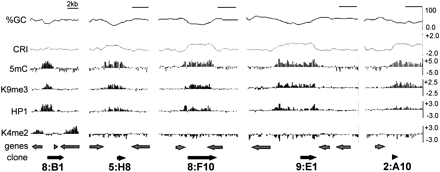
Chromatin modification profile of previously identified methylated regions. Data from five representative control regions are shown. A scale bar indicating 2 kb is shown at the top right of each plot. The base composition (%GC) and a composite RIP index (CRI) of each region, plotted as the moving average for 500 bp windows with 100 bp steps, are shown at the top of each plot. Enrichment values for MeDIP and ChIP-chip experiments are shown as log2 values (y-axis) for immunoprecipitation experiments with antibodies to 5mC, H3K9me3, green fluorescent protein for HP1-GFP (HP1) and H3K4me2 for each region. The positions of predicted open reading frames (genes) and the previously identified methylated DNA clones, with their identification numbers, are shown at the bottom.











