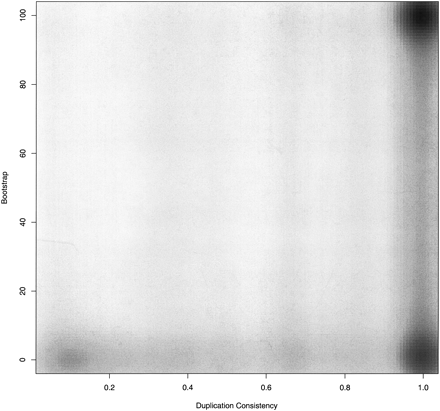
Figure 3.
A scatter plot of the duplication consistency score (x-axis) compared to the bootstrap value of duplication nodes (y-axis). Because of the large number of values, the density of points is shown using the smoothScatter kernel-based density function in R.











