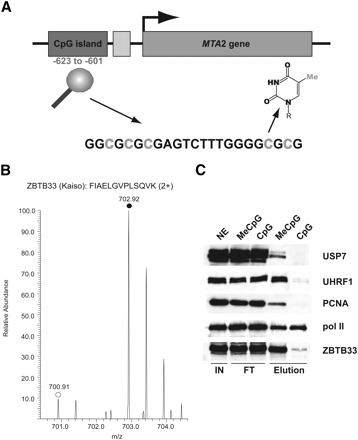
Methyl-CpG specific binding partners can be discriminated by SILAC analysis. (A) Scheme of the CpG island in the upstream region of the human MTA2 gene that is located at position −623 to −601 relative to the transcription start site (arrowhead). The cytosine residues highlighted in gray were either fully methylated (MeCpG) or not methylated (CpG) in the SILAC experiment. The structure of 5-methyl cytosine is shown. (B) Representative peptide mass spectrum demonstrating the preferential binding of the methyl-CpG binding protein ZBTB33 (Kaiso), which was known to interact with the fully methylated MTA2 CpG island. The MS spectrum of a labeled (monoisotopic peak marked with filled circle) and unlabeled (monoisotopic peak marked with open circle) tryptic ZBTB33 peptide acquired is shown. (C) Western blot analysis (same conditions as in Fig. 2C) of selected specific interaction partners recapitulates results from the SILAC analysis (Table 2).











