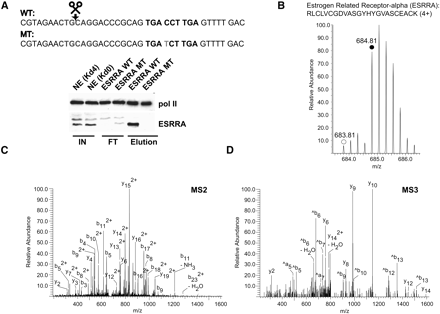
The orphan nuclear receptor ESRRA (ERRalpha) exhibits specific binding to its bioinformatically predicted binding sequence. (A) Western blot analysis (same conditions as in Fig. 2C) confirms the specific interaction of ESRRA as revealed by the SILAC experiment (B–D) (Table 1). Sequences of the DNA baits employed are depicted. The predicted ESRRA binding site derived from an autoregulatory motif of the ESRRA gene (Mootha et al. 2004) is shown in boldface type whereas the point mutation for the control column is highlighted in gray. (B) Representative peptide mass spectrum demonstrating the specific binding of ESRRA. The MS spectrum of a quadruply charged labeled (monoisotopic peak marked with solid circle) and unlabeled (monoisotopic peak marked with open circle) tryptic ESRRA peptide acquired in the ICR cell of the LTQ-FT (0.3 ppm mass deviation after recalibration) is shown. (C) MS/MS (MS2) fragmentation spectrum that identifies the peptide shown in B. The precursor ion (m/z 684.81) was fragmented in the linear ion trap (LTQ part) of the mass spectrometer to obtain sequence information. (D) MS/MS/MS (MS3) spectrum of the y 15(2+) ion present in the MS2 experiment (C) further confirming the identification of the peptide shown in B.











