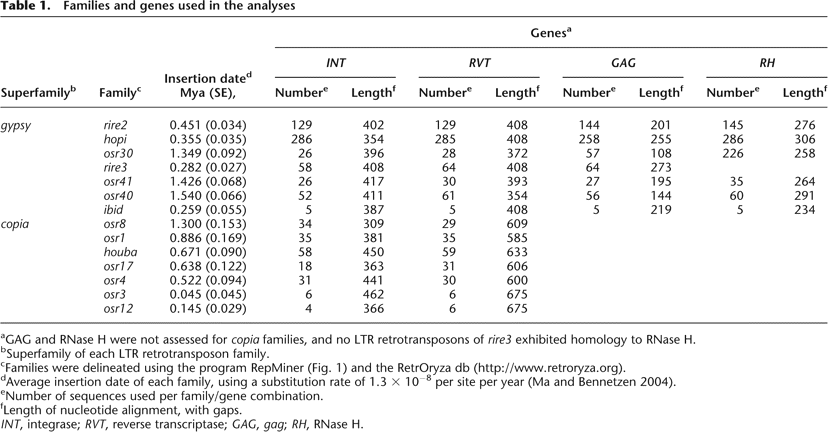
Families and genes used in the analyses
Click on table to view larger version.
-
aGAG and RNase H were not assessed for copia families, and no LTR retrotransposons of rire3 exhibited homology to RNase H.
-
bSuperfamily of each LTR retrotransposon family.
-
cFamilies were delineated using the program RepMiner (Fig. 1) and the RetrOryza db (http://www.retroryza.org).
-
dAverage insertion date of each family, using a substitution rate of 1.3 × 10−8 per site per year (Ma and Bennetzen 2004).
-
eNumber of sequences used per family/gene combination.
-
fLength of nucleotide alignment, with gaps.
-
INT, integrase; RVT, reverse transcriptase; GAG, gag; RH, RNase H.











