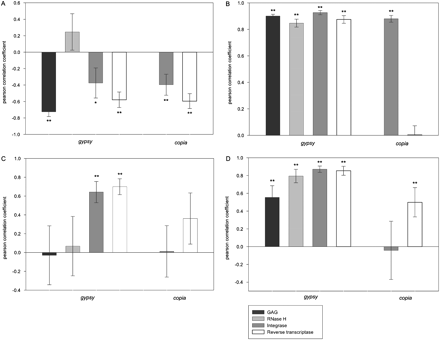
The strength and direction of the correlations between the average insertion date of the LTR retrotransposon families in rice and Ts:Tv (A), the proportion of premature stop codons within each family (B), πs (C), and πn/πs (D), presented for each gene analyzed of each superfamily. Standard errors around the mean are represented by 95% confidence intervals, estimated from the jackknife procedure. (*) The correlation was significant at the P < 0.05 level; (**) significance at the P < 0.005 level after Bonferroni corrections. (Black bars) gag; (light gray bars) RNase H; (dark gray bars) integrase; (open bars) reverse transcriptase.











