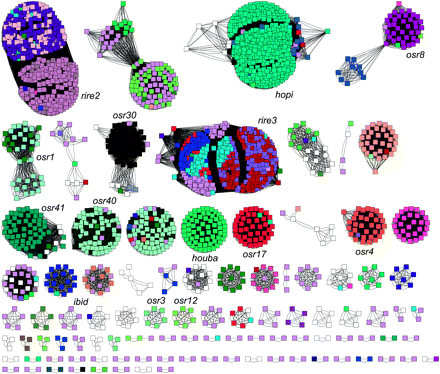
The LTR retrotransposon families of rice. An all-by-all BLASTN (e−10) was performed using the 5′ LTR sequence of each element from our database of 2226 LTR retrotransposons. The program RepMiner (J. Estill) was then used to format the results of this BLAST for graphic display by the program CytoScape (v. 2.5.1). The 5′ LTR of each element is illustrated as a single square node, while blast hits are shown as lines connecting these nodes. Each individual cluster of connected nodes may be considered a family according to a connected components interpretation of family affinity. The colors of individual nodes represent the family name assignment from a BLASTN (e−10) to the RetrOryza database (Chaparro et al. 2007). Families used in the selection analyses are labeled.











