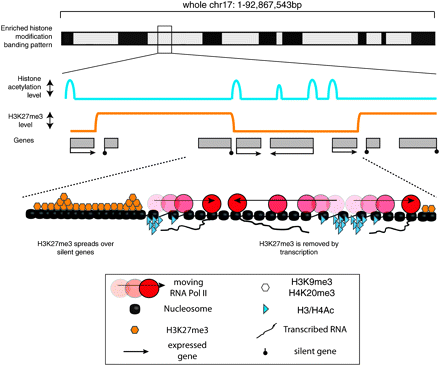
Summary of H3K27me3 distribution along an autosomal chromosome. (Top) Predicted Giemsa banding pattern in metaphase chr 17 predicted from the data obtained in Figure 6 that is based on a nonsynchronized, mainly interphase cell population. (Middle) Schematic showing enlarged detail of the histone modification profile in regions with a high gene and SINE density that are enriched for repressive H3K27me3 and multiple active histone modifications. At a 100 bp high resolution level, repressive H3K27me3 and active H3Ac/H4Ac histone modifications are mutually exclusive. Expressed genes (black boxes) are characterized by active histone modifications at promoters (blue line) or by H3K36me3 throughout the gene body (not shown). In contrast, silent genes (gray boxes) and nontranscribed intergenic regions are covered by large BLOCs of H3K27me3 (orange line). BLOCs have sharp boundaries that are immediately flanked by transcripts from expressed genes. (Bottom) Enlarged detail showing sites of initial H3K27me3 deposition with the highest levels of H3K27me3 (piled orange hexagons). H3K27me3 could then spread in an unknown manner, along the chromosome, but is excluded or erased by RNA Pol II (red circles) or blocked by boundary elements marked by active histone modifications (blue triangles).











