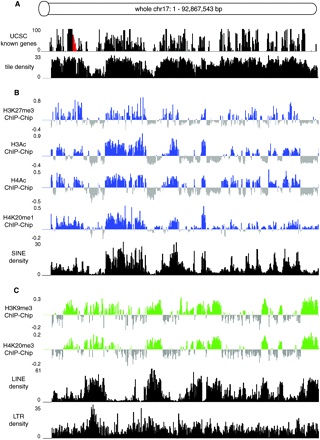
Histone modifications identify two types of chromosome bands that correlate with gene and repeat density. (A) Gene density (http://genome.ucsc.edu) and probe tiling density (repeat-masked, see Methods for details) on the whole chr 17 tiling array chip, were analyzed in 200 kb nonoverlapping windows. The red area marks the imprinted Igf2r cluster (shown in Fig. 4) that contains three small H3K9me3/H4K20me3 peaks and lacks H3K27me3 (Regha et al. 2007). (B) Cumulative ChIP-chip profiles (log2 ChIP/Input hybridization signal means from MEFF and MEFB1 cells, Supplemental Table 1) for H3K27me3, H3Ac, H4Ac, H4K20me1 (blue bars), and SINE density (black bars). Each vertical bar indicates an average signal from 200 kb nonoverlapping windows. (C) Cumulative ChIP-chip profiles for H3K9me3 and H4K20me3 (green bars) and LINEs and LTRs (black bars). Details as in (B).











