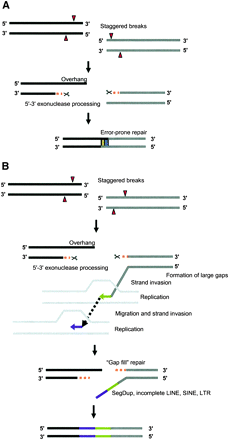
Models for human–NLE gibbon rearrangements. (A) An error-prone repair mechanism for smaller breakpoint intervals (<20 bp). DNA strands from two chromosomes (black and gray bars) are shown. Staggered double-strand breaks are processed by 5′–3′ exonuclease, creating overhangs. These overhangs are filled by an error-prone repair mechanism, creating shorter insertions. (B) “Gap-fill” model for larger breakpoint intervals. Large gaps are generated by double-strand breaks (due to possible collapsed or stalled replication forks) at rearrangement sites. These staggered breaks are processed by exonucleases to generate long 3′ overhangs. Replication is initiated by strand invasion to repair the gap. However, likely due to low processivity of the replication-dependent repair process (McVey et al. 2004), only smaller-length sequence stretches are synthesized. Consequently, a series of strand invasion, replication, and uncoupling of the replication machinery is necessary to fill the large gap. Thus, a less-efficient replication-based repair process generates a mosaic of incomplete repetitive elements at the larger breakpoint intervals.











