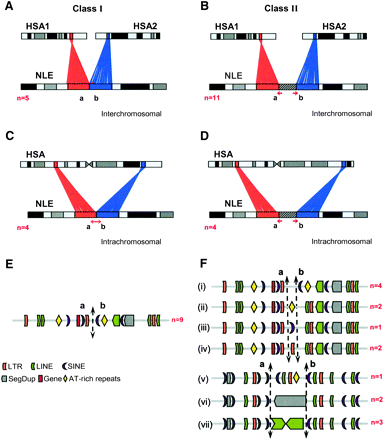
Class I and class II breakpoints. The schematic shows the types of rearrangements identified by high-resolution sequence analysis: Class I and class II breakpoints causing inter- (A,B) or intrachromosomal (C,D) rearrangements are shown. Based on the sequence context, the number (n) of different human–gibbon breakpoints identified from both categories (E,F) are also shown. Note that the class II breakpoints contain: (i) nonrepeat sequences, (ii) AT-rich repeats, (iii) SINEs (AluY element), (iv) LTR insertions, (v) a “hodgepodge” of repeats, (vi) segmental duplications, and (vii) LINE-1 elements. The diagram is not to scale.











