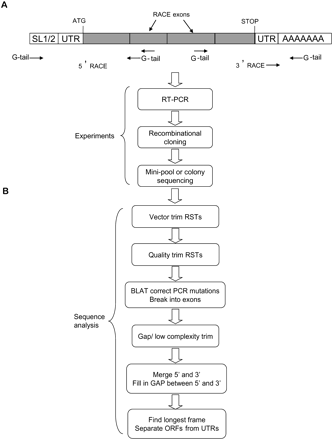
Experimental and computational RACE pipelines. (A) Experimental strategy begins with reverse transcription of messenger RNA (with a tailed dT24 for 3′-RACE) to generate a complementary DNA strand used as template to generate the first set of RACE products. These amplicons are then reamplified with nested internal primers to generate the final RACE products, which are cloned, then sequenced. (B) Computational pipeline begins with vector and quality trimming of the reads. RACE sequences are aligned by BLAT against the C. elegans genome then parsed as genome (exon) blocks. RACE sequences are then replaced by matching genomic sequences to correct sporadic sequencing or PCR errors. If an exon is of low complexity or has regions that cannot be aligned to the genome, the exon, together with subsequent exons (if present), is trimmed. For short transcripts, 5′ RACE Sequence Tags (RSTs) and 3′ RSTs can be readily assembled to generate full transcripts. For longer transcripts, the gaps of nonoverlapping 5′ RSTs and 3′ RSTs are filled by sequences from WormBase transcript models.











