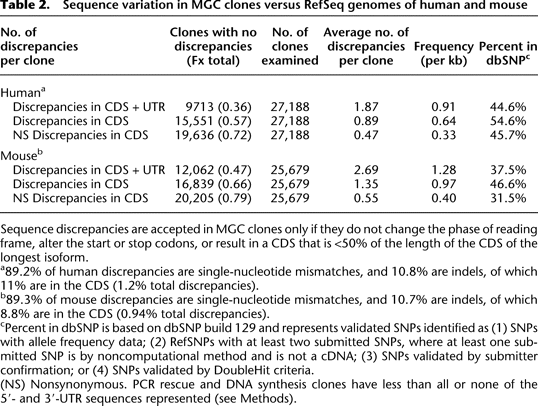
-
Sequence discrepancies are accepted in MGC clones only if they do not change the phase of reading frame, alter the start or stop codons, or result in a CDS that is <50% of the length of the CDS of the longest isoform.
-
a89.2% of human discrepancies are single-nucleotide mismatches, and 10.8% are indels, of which 11% are in the CDS (1.2% total discrepancies).
-
b89.3% of mouse discrepancies are single-nucleotide mismatches, and 10.7% are indels, of which 8.8% are in the CDS (0.94% total discrepancies).
-
cPercent in dbSNP is based on dbSNP build 129 and represents validated SNPs identified as (1) SNPs with allele frequency data; (2) RefSNPs with at least two submitted SNPs, where at least one submitted SNP is by noncomputational method and is not a cDNA; (3) SNPs validated by submitter confirmation; or (4) SNPs validated by DoubleHit criteria.
-
(NS) Nonsynonymous. PCR rescue and DNA synthesis clones have less than all or none of the 5′- and 3′-UTR sequences represented (see Methods).











