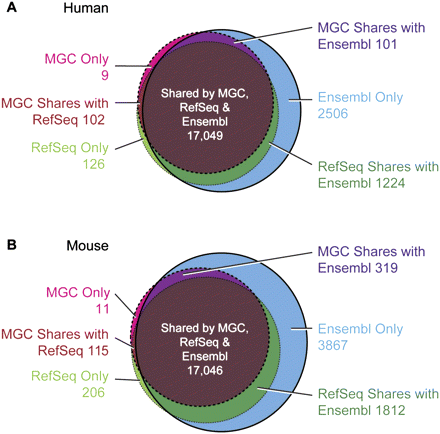
Venn diagram comparing the number of loci containing protein-coding genes from MGC, RefSeq, and Ensembl. (A) Human; (B) mouse. The loci were computed by clustering transcripts from all three gene sets based on the overlap of the genomic location of the CDS portion of the exons. When a transcript is not uniquely mapped to the genome, the clusters for all mappings of that transcript were combined and counted as one locus. For human, this resulted in 17,239 loci containing MGC clones, 18,494 loci with RefSeq mRNAs (Pruitt et al. 2009b), and 20,856 Ensembl gene loci (Hubbard et al. 2002). Mouse had 17,455 loci with MGC clones, 19,064 loci with RefSeq mRNAs, and 23,087 Ensembl gene loci. Genes counted as shared between any two gene sets exclude genes in the third set. BLAT (Kent 2002) alignments of MGC clones and RefSeq mRNAs (NM accessions) obtained from the UCSC Genome Browser database (Karolchik et al. 2008) for human genome assembly 36.1 and mouse assembly 37, and Ensembl Release 52 were used in the analysis. Genomic loci serve as an estimate of the number of genes in these data sets. The counts vary from those seen in Table 1, owing to the different method of computation.











