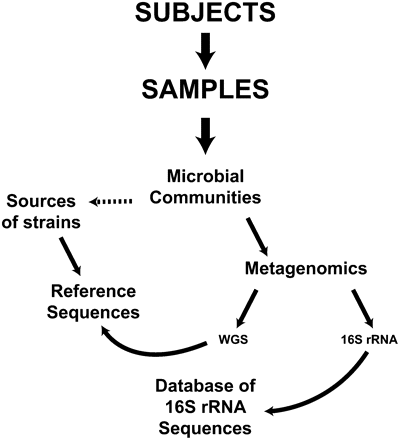
Scheme for studying the “normal” human microbiome. Subjects are recruited, and samples are collected. The microbial communities in the samples are analyzed using metagenomic sequencing approaches. 16S rRNA sequences are compared to databases of 16S rRNA sequences, while whole-genome shotgun (WGS) sequences are compared to existing reference strains, many of which will have been sequenced under the HMP. The microbial communities from the donors can also serve as sources of strains that will be sequenced to increase the number of known reference sequences.











