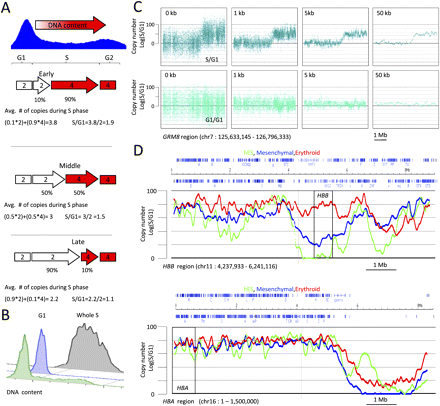
TimEX. (A) Principle of the technique. Copy number of DNA in sorted S-phase cells compared with sorted G1 cells can be used as a surrogate measurement for the timing of replication (see text). (B) Typical pre- and post-sort DNA content profiles of cycling basophilic erythroblasts detected by staining with propidium iodide. Green profile, pre-sort DNA content profiles; blue and gray, respectively, G1 and S post-sort profiles. (C) Scatterplots illustrating smoothing by Gaussian convolution. Top and bottom panels are, respectively, scatterplots of S/G1 and control G1/G1 ratios for the 8-mb GRM8 region on chr 7 (see Supplemental Table S1). (X-axis) Genomic position; (y-axis) normalized S/G1 or G1/G1 ratio. The left panels illustrate the results without any smoothing; the three panels on the right show the same data smoothed by Gaussian convolution of sigma equal to 1, 5, or 50 kb. As expected, the G1/G1 ratio is flat, while the S/G1 ratio varies. High S/G1 ratios indicate regions that replicate early in S phase, low S/G1 ratios regions that replicate late in S phase. (D) Comparison of the timing of replication in hESCs and in mesenchymal and erythroid cells derived from hESCs. The scatterplots are as above. The red, green, and blue curves, respectively, represent the TimEX profiles of the three cell types. A total of 8-Mb regions containing the beta hemoglobin (HBB) and the alpha hemoglobin (HBA) are shown. All of the other regions present in the arrays are shown in Supplemental Figure S2. The profiles in the three cell types are different, but the overall shape of the curves and the slopes of the transition regions are similar, suggesting that the underlying molecular mechanisms are the same. Differences between cell types are particularly evident in gene-poor regions.











