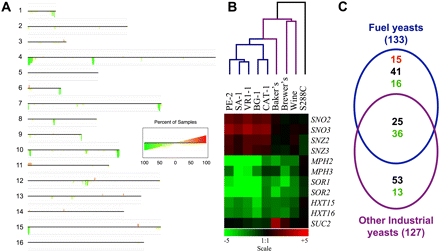
Changes in gene copy number shared by the fuel ethanol yeast strains. (A) A consensus karyoscope plot pooling the results of all the fuel ethanol strains relative to the lab strain S288C was generated by the program CGH-Miner (Wang et al. 2005). The plot displays statistically significantly altered copy number variations along each chromosome, and the chromosomes are shown in numerical order from top to bottom. (Red bars above the chromosome) significantly amplified regions, (green bars below the chromosome) significantly depleted regions; the height of the bars indicates in how many of the strains the significant observation was made (see reference scale within the figure). (B) Clustering dendogram of the entire genome microarray CGH data for the selected fuel yeast strains, together with the same data obtained with a baker's, brewer's, wine, and the lab strain S288C (self–self hybridization) (top), and relative gene copy number of selected yeast genes among the different strains (bottom). A scale of relative gene copy number is shown at the bottom. (C) Venn diagram of number of altered gene copy number variations of the selected fuel-ethanol yeast strains that were shared (or not) with a group of nine other industrial yeast strains (four wine, four baker's, and one ale brewing strain). (Red) Amplified genes, (black) polymorphic loci, (green) depleted genes (see Methods).











