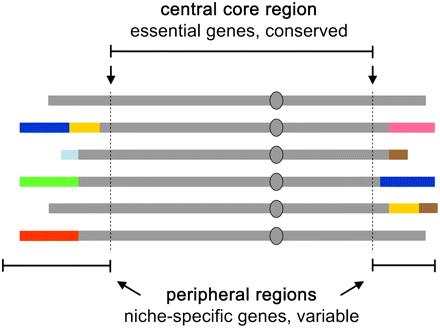
Structural diversity in S. cerevisiae: rigid and plastic domains of the genome. The model depicts a set of homologs of a hypothetical chromosome in several unrelated S. cerevisiae strains. The top line (continuous gray) depicts the structural configuration of this chromosome in the first sequenced strain (i.e., S288c; reference), whereas the chromosomes shown below represent the rearrangements found in other strains. The diverged structural configurations (colors) in the peripheral regions harbor genes that are not required for viability, but that may contribute to fitness in specific environments. The entire set shares structural conservation in the central core region (delimited by the most distal essential genes, arrows); therefore, meiotic crossovers between the unrelated haplotypes can generate new combinations, while remaining compatible with haploid viability.











