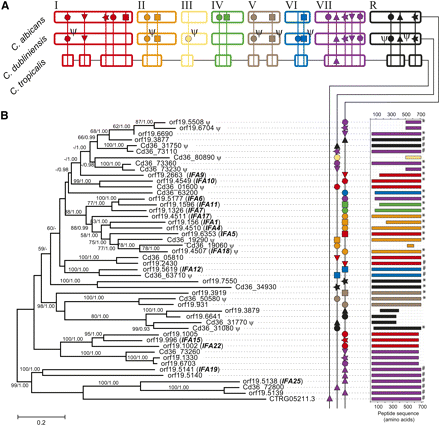
Phylogeny and comparative genomics of the IFA gene family in Candida spp. (A) Cartoon of genomic distribution (note: not precise karyotypes). Pseudogenes are denoted by Ψ. (B) Maximum likelihood phylogeny of IFA protein sequences in C. albicans (prefixed orf19.), C. dubliniensis (prefixed Cd36_), and C. tropicalis (CTRG05211.3). Branch lengths are proportional to evolutionary change and measured in substitutions/site. This topology was concordant with an alternative Bayesian consensus tree. Dotted lines link gene IDs to a cartoon of a multiple sequence alignment. C termini are labeled according to amino acid composition: (#) long, acidic-type; (*) a shorter, nonacidic-type C-terminal. No label indicates that neither type is present or the C terminus is absent.











