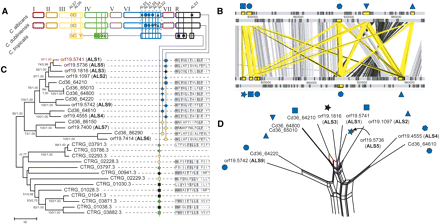
Comparative genomics and phylogeny of agglutinin-like sequences (ALS) in Candida spp. (A) A cartoon showing genomic distribution of ALS loci in C. albicans, C. dubliniensis, and C. tropicalis. The positions of loci are indicated by vertical lines and the presence of a gene(s) is represented by a unique symbol (which is also applied to positional orthologs in other species). A dashed line indicates the region of chromosome 6 expanded in panel C. Note that the chromosomes are drawn based on the C. albicans karyotype, with C. dubliniensis and C. tropicalis pseudochromosomes drawn for comparison, reflecting their conserved gene order, but not precise karyotypes. (B) Maximum likelihood phylogeny of ALS N-terminal nucleotide sequences in C. albicans (for which gene names ALS1-9 are given), C. dubliniensis (prefixed Cd36_) and C. tropicalis (prefixed CTRG_). Branch lengths are proportional to evolutionary change and measured in substitutions/site. This topology was concordant with an alternative Bayesian consensus tree; each node is attended by nonparametric bootstrap values and posterior probabilities from likelihood and Bayesian analyses respectively. Terminal nodes are labeled with gene IDs and locus symbols (as used in A), and then to the 14 amino acid sequence of the C terminus (where available). (C) ACT representation of ALS loci on chromosome 6 in C. dubliniensis (top) and C. albicans (bottom). ALS genes are shaded yellow and marked as in A. Significant TBLASTX hits between genes are represented by vertical bars (gray, sense; black, anti-sense; yellow, ALS). (D) Phylogenetic network showing a consensus of all possible relationships among chromosome 6 genes simultaneously. Splits supporting monophyly of Cd36_64210 and ALS1 are shown in red.











