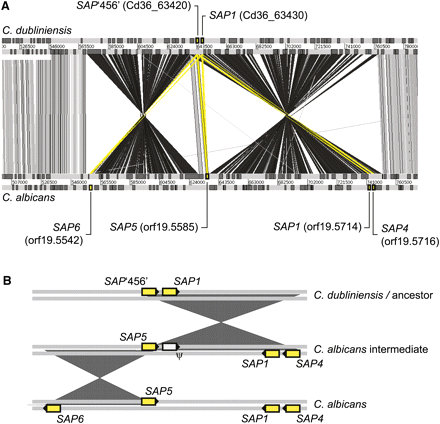
Segmental inversion and the evolution of secreted aspartyl proteinases (SAP). (A) Comparison of SAP genes on chromosome 6 (0.5–0.8 Mb) in C. albicans and C. dubliniensis generated using the Artemis Comparison Tool (ACT); DNA strands are represented by horizontal gray bars (scale in base pairs); genes are indicated by darker boxes. SAP genes are shaded yellow and linked to systematic IDs. Significant TBLASTX hits between genes are shown by vertical bars (gray, sense; black, anti-sense; yellow, SAPs). (B) A cartoon of the hypothesis that two segmental inversions created two additional SAP loci in C. albicans. The first inversion duplicated the original tandem gene pair (“C. dubliniensis/ancestor”) creating paralogs of SAP1 and SAP‘456’ in the opposite orientation (“C. albicans intermediate”); this is consistent with SAP1 and SAP4 in C. albicans and demands that the original SAP1 copy was subsequently lost (ψ). The second inversion duplicated the remaining SAP‘456’ copy (i.e., SAP5) to create a single gene in the opposite orientation (i.e., SAP6).











