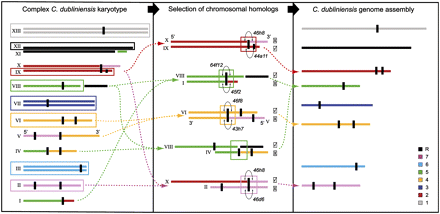
The C. dubliniensis complex in vivo karyotype and genome sequence. The 14.6-Mb C. dubliniensis genome (left panel, redrawn with permission from Magee et al. 2008) has a complex karyotype of 10 haploid and three diploid chromosomes (Roman numerals), showing multiple chromosomal rearrangements relative to C. albicans. Vertical black marks denote MRS regions. In order to avoid redundancy in the assembly and to maximize comparability with C. albicans, the sequence was assembled into eight “pseudochromosomes” (right panel), which are numbered according to the karyotype of C. albicans strain SC5314 (Arabic numerals) and color-coded after C. albicans in order to illustrate the extent of chromosomal rearrangement in vivo. Note that each pseudochromosome in the right panel is a haploid consensus of the distinct chromosomal homologs apparent in the left panel. Components of the in vivo karyotype selected for the genome sequence are boxed. C. dubliniensis chromosomal homologs often adopted two alternative configurations, relative to the C. albicans karyotype. The middle panel illustrates the selection process by which one haploid chromosome was chosen over its homolog, to reproduce the C. albicans karyotype. Tick boxes indicate which chromosomal configuration was selected in the final genome representation (as submitted to the EMBL database). BAC clones were sequenced that bridged the two distinct chromosomal configurations to confirm that both existed (BAC clone identifiers are noted in italics where appropriate). Note that chromosome V is inverted.











