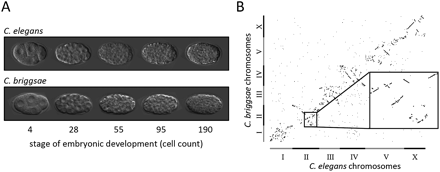
C. elegans and C. briggsae development is indistinguishable despite a large genetic distance. (A) Micrographs showing C. elegans and C. briggsae embryos at the embryonic developmental stages examined in this study. The cell division patterns are remarkably similar in terms of cell deaths, relative division times, and cell locations within the embryo (Zhao et al. 2008). (B) Dot plot showing the genomic locations of one-to-one orthologs between the C. elegans and C. briggsae genomes, indicating numerous genomic rearrangements.











