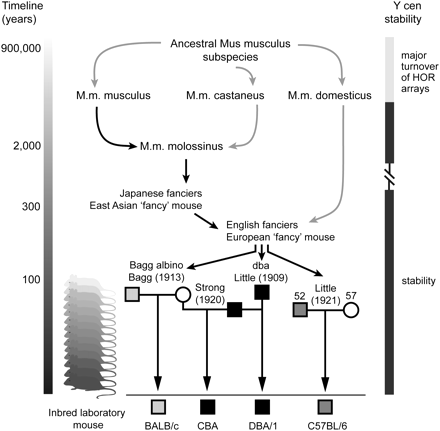
Origins of inbred laboratory mice and evidence for sequence stability at the mouse Y centromere. Present day Mus musculus subspecies diverged from a common ancestor around 900,000 yr ago. Recent extensive hybridization in Japan between M.m. musculus and M.m. castaneus subspecies has given rise to the M.m. molossinus hybrid, which carries the M.m. musculus Y chromosome and M.m. castaneus mitochondrial DNA (M.m. musculus Y transmission depicted by black arrows). Many classical inbred laboratory mouse strains have a M.m. musculus (molossinus) Y chromosome following inbreeding of pet “fancy mice” of East Asian and European ancestry by mouse geneticists (e.g., Bagg, Strong, Little) early last century. The origin of four inbred laboratory strains carrying three phylogenetically distinct molossinus Y chromosomes is shown: light gray box, male BALB/c; black box; male DBA/1 and CBA; dark gray box, male C57BL/6J. Strains DBA/1 and CBA carry the same ancestral Y chromosome inherited from a dba male (the first inbred strain established by Clarence C. Little in 1909; cascading mice at left representing clonal inheritance). At the resolution of PFGE, these strains carry an identical 90-kb Ymin array length, suggesting stability of the same Y centromere array over at least 90 yr and 167 (DBA/1) and 175 (CBA) inbred generations. Strains C57BL/6J, BALB/c, and 129 (data not shown) also show 90-kb array lengths, increasing total recorded inbred generations to 734 without a detectable change to the Y centromere array length. These data suggest changes to the array occur below the resolution of PFGE (possibly in a small, stepwise fashion) or they occur very rarely. Despite this apparent stability, rapid divergence of Ymin DNA sequence and HOR unit structure is evident at the Y centromeres of M.m. molossinus and M.m. domesticus mice, driven by a major turnover of the Ymin HOR arrays (see main text for details).











