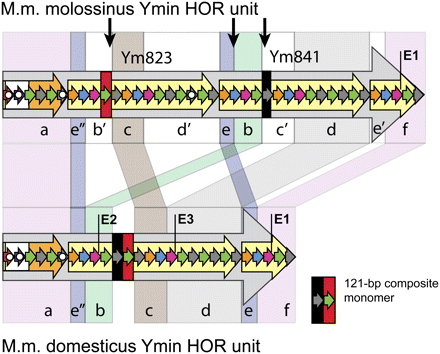
Comparative sequence analysis of M.m. molossinus (2.3 kb) and M.m. domesticus (1.6 kb) Ymin HOR units. Regions of highest sequence homology are grouped (a–f) and shaded accordingly. Landmark EcoRI restriction sites are labeled E1–E3; monomers harboring deletions or other rearrangements are highlighted by solid white circles (see Fig. 1F for full description). The M.m. domesticus HOR unit contains a 121-bp composite monomer that is absent from the equivalent regions of the M.m. molossinus Ym823 and Ym841 subunit repeats. Monomers with highest sequence identity to this composite monomer are highlighted in the molossinus HOR using red and black bars, and border regions of putative unequal exchange (vertical black arrows). Differences between the two HOR unit structures can be explained by a series of unequal sister chromatid exchange events occurring in the ancestral HOR arrays, followed by amplification and fixation of the present day canonical HOR units in each subspecies. At least three unequal exchange events are required to reconcile key differences between the two HOR units (for proposed model of unequal exchange, see Supplemental Fig. S8).











