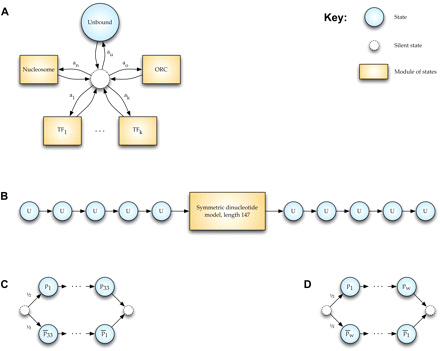
State transition diagram for Boltzmann chain implementation in COMPETE. Blue circular nodes are states and correspond to single nucleotides. Dashed circular nodes are silent states because they do not correspond to any nucleotide. Orange rectangular nodes are modules of states and correspond to a sequence of nucleotides of some length. Edges represent probabilistic transitions between states, with transition probabilities labeled; to reduce clutter, if a node has only one outbound edge, it is unlabeled (the probability is exactly 1). (A) At the highest level, the model represents each position in the genome either as being in an unbound state or as being bound by one of any number of DNA binding proteins or protein complexes. For instance, we might choose to model the genome as being bound by nucleosomes, the origin recognition complex (ORC), and k different transcription factors (TFs). Each DBF is represented by a module of an appropriate length. Transition probabilities from the central silent state are proportional to concentrations of the respective DBFs. (B) The nucleosome module consists of a symmetric dinucleotide model of length 147 flanked by 5 unbound nucleotides on either end to enforce a spacing between nucleosomes of at least length 10. To reduce clutter, we do not represent the states within the central module in this figure. (C) The ORC module consists of a PSSM-based EACS+B1 motif of length 33 (Xu et al. 2006) in either a forward or reverse-complement orientation (the origin might be on either strand, and we assume that each occurs with probability 1/2). (D) A TF module consists of a PSSM-based motif of length w, a value that varies for each TF. Because a TF can bind to either strand, the w nucleotides arise either from the PSSM or from its reverse complement, each with probability 1/2.











