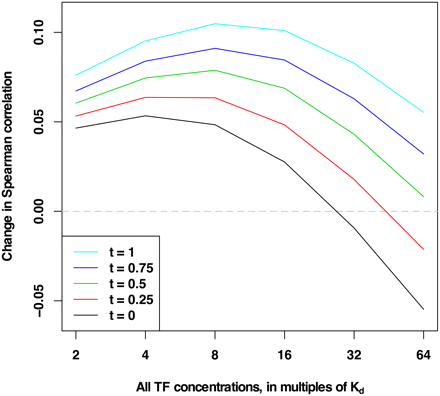
Change in Spearman correlation with and without TFs. DBF competition improves nucleosome positioning predictions genome-wide. Enriched and depleted regions across the genome are extracted from the in vivo experimental map of Kaplan et al. (2009) using different stringency thresholds t, with all positions in enriched regions having experimental nucleosome occupancy greater than t and all positions in depleted regions having experimental nucleosome occupancy less than −t. The Spearman correlation is computed between the experimentally measured nucleosome occupancy of the regions and the occupancy predicted by COMPETE using nucleosome models with and without all 89 TFs of Zhu et al. (2009). The TF concentrations are all set to the same multiples of their respective Kds. Each point corresponds to a pair of whole-genome analyses, with and without all 89 TFs, totaling 12 decodings of the genome and 60 individual analyses. The changes between Spearman correlations of experimental data and COMPETE predictions with and without nucleosomes are shown here, where positive values signify improvement. Inclusion of TFs is clearly beneficial to nucleosome positioning across TF concentrations. This effect ranges in extent and is demonstrated for various TF concentrations and various thresholds t. Raising TF concentrations to high levels imposes deleterious effects on nucleosome positioning, likely due to diminished nucleosome occupancy in competition with many different highly concentrated TFs. When TF concentrations are kept below these high levels, TF inclusion aids significantly in nucleosome binding predictions. Values of all points are given in Supplemental Table S2.











