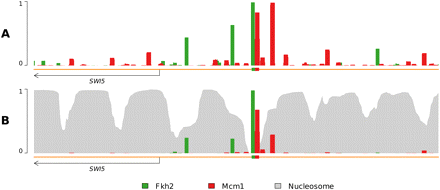
Nucleosome binding attenuates nonfunctional transcription factor binding. (A) A profile using a model of only TFs and DNA yields many (presumably nonfunctional) binding predictions throughout the genome. The binding of Fkh2 and Mcm1 is significant even away from their annotated binding sites; Mcm1 even displays an upstream (to the right) binding occurrence stronger than at its annotated site. However, Fkh2 and Mcm1 form a ternary complex with DNA and are known to bind adjacently to one another, making the observation of this additional, stronger, binding occurrence somewhat curious. (B) Addition of nucleosomes yields a dramatically altered TF occupancy profile. Fkh2 and Mcm1 binding is notably diminished at sites other than annotated binding sites, though not entirely eliminated. Additionally, the strong upstream Mcm1 binding event occurs at a much-reduced level, now approximately half as frequently as that of the annotated Mcm1 binding site.











