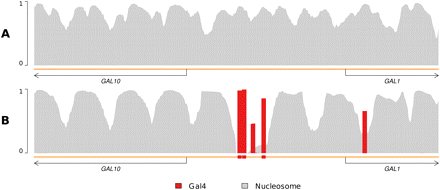
Transcription factor binding can stabilize nucleosomes. Shown are DBF occupancy profiles. The x-axis is the genome position, and the y-axis is the probability of occupancy. Plots with each legend color indicate the probability of the respective DBF occupying each position. The orange line underneath each plot represents the genome; oriented arrows demarcate coding regions of genes according to SGD (Cherry et al. 1998); colored boxes denote strong TF binding site matches according to MacIsaac et al. (2006). (A) Including only nucleosomes and unbound genomic sequence yields an occupancy profile similar to existing nucleosome-only positioning models like that of Segal et al. (2006). In this genomic region, the degenerate nature of nucleosome sequence preferences leads to significant uncertainty of nucleosome positioning. (B) Addition of Gal4 yields a strikingly different nucleosome occupancy profile, revealing stable nucleosome positioning. As Gal4 strongly binds its annotated binding sites, nucleosomes are outcompeted at those locations, and “boundaries” are established. These boundaries reduce the feasibility of nucleosome translation, so nucleosome positions coalesce into more stable locations in response.











