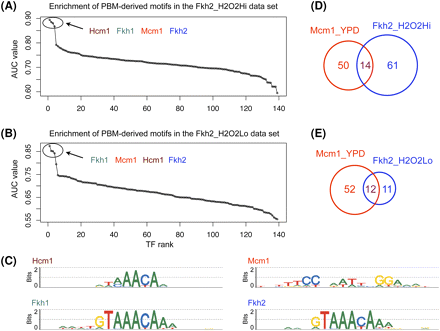
High-scoring motifs in the Fkh2_H2O2Hi and Fkh2_H2O2Lo ChIP-chip data sets. (A,B) AUC values for the 139 PBM-derived motifs in the two data sets. The x-axes show the TF ranks, computed as in Figure 1C. (C) Motifs significantly enriched in the two data sets. The DNA binding motif of Fkh2 was correctly identified as one of the significantly enriched motifs. In addition to Fkh2, the Hcm1, Fkh1, and Mcm1 motifs are also highly enriched. The Hcm1 and Fkh1 motifs are similar to the Fkh2 motif. Mcm1 is known to bind cooperatively with Fkh2 (Hollenhorst et al. 2001). (D,E) Venn diagrams showing the overlaps between the sets of probes bound by Fhk2 and Mcm1 in different environmental conditions.











