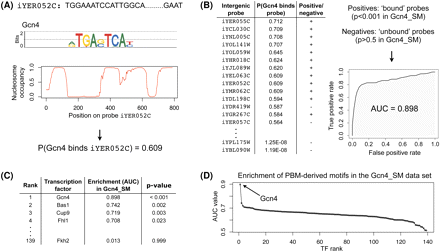
Identification of highly enriched motifs in a ChIP-chip data set. We proceed in four steps: (A) For each TF with a PBM-derived motif (here, Gcn4) and each intergenic probe (here, iYER052c), we compute the probability that the TF binds that probe, as described in the Methods section. (B) For each TF (here, Gcn4) we rank all intergenic probes in decreasing order of the binding probability and then compute the enrichment of the motif in a ChIP-chip data set (here, Gcn4_SM) according to AUC. To calculate the AUC statistic, we defined the positive and negative sets to be the sets of intergenic regions with ChIP-chip P-values < 0.001 and >0.5, respectively, as calculated by Harbison et al. (2004). (C) For each ChIP-chip data set (here, Gcn4_SM), we ranked all TFs in decreasing order of their motif's AUC value. (D) We determine the significantly enriched motif(s) (here, Gcn4).











