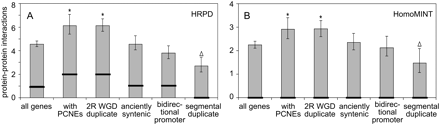
Retained WGD duplicates are overconnected and segmental duplicates are underconnected. The degree of protein–protein interaction (PPI) connectivity was estimated for mouse gene sets with different genome evolutionary characteristics using two different PPI databases, HPRD (A) and HomoMINT (B). Bar heights indicate the mean connectivity for each gene set. Each mean is bounded by a bootstrap-based 95% confidence interval. Median connectivity is indicated as a bold line, and the difference between the median and mean gives some indication of the right-skew of the connectivity distribution for each gene set. Genes with PCNEs or that are retained WGD duplicates have significantly more PPI connections than the genome mean (asterisks), while genes that are the result of segmental duplication have significantly fewer connections (triangles). Significance was assessed by a Bonferroni-corrected permutation test, α = 0.05.











