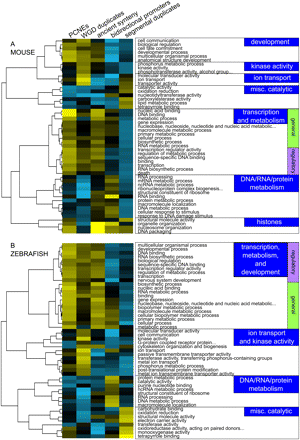
Different gene functions are associated with distinct patterns of genome evolution. Hierarchical clustering of gene functions (rows) and five genome evolutionary processes (columns): linkage to PCNEs, retention of duplicate genes after the early vertebrate WGD events, synteny conservation, bidirectional promoter, and segmental duplication. The gene functions are derived from the biological process and molecular function namespaces of the Gene Ontology; the 50 classes with the strongest associations are shown, using data from the mouse (A) and zebrafish (B) genomes. Yellow colors indicate that genes annotated with the GO term are more strongly associated with the genome evolutionary process than the genome-wide average; blue colors indicate less than the genome-wide average. The clustering procedure divides GO terms into groups that represent broad cellular processes (labels on the right), each of which is associated with a distinct pattern of genome evolution. All olfactory receptor genes were removed from the data prior to this analysis.











