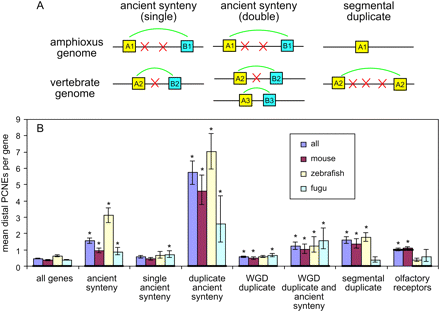
Genes with different synteny and duplication patterns have different PCNE amounts. (A) Illustration of our synteny and segmental duplication definitions. (B) The mean number of extragenic PCNEs linked to different synteny and duplication related gene sets. Error bars, bootstrap-based 95% confidence intervals on the mean estimates. Asterisks denote PCNE enrichments that are significant at α = 0.05 by a Bonferroni-corrected permutation test. Bold lines show the population medians, which are zero in all cases except for the olfactory receptor genes. This serves to emphasize the fact that in all populations the majority of genes have no PCNEs, and the observed differences between the gene sets, while significant, are generally the result of changes in PCNE number around a subset of genes.











