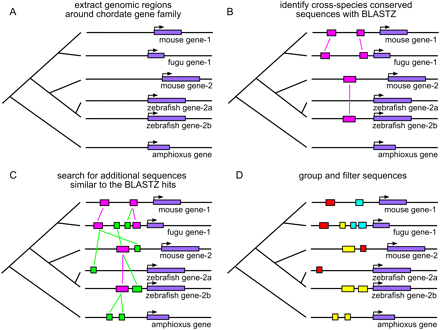
Overview of our method for identifying PCNEs around a hypothetical chordate gene family. (A) Genes related to a single ancestral chordate gene are identified, and the genomic sequences surrounding these genes are extracted, extending out 100 kb on either side of the genes. (B) A fast search algorithm (BLASTZ) is used to identify conserved noncoding sequences in cross-species comparisons. (C) A slower, more sensitive algorithm (CHAOS) searches for additional noncoding sequences similar to the BLASTZ elements. (D) The resulting conserved sequences are grouped into loose families and filtered to remove low-confidence sequences and any remaining coding sequences.











