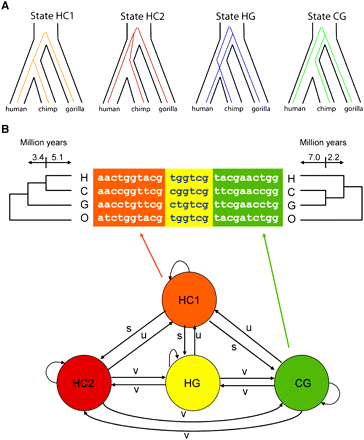
(A) The four genealogy types associated with the states of the hidden Markov model of Hobolth et al. (2007). State HC1 describes the case in which the human/chimpanzee coalescence occurs subsequent to the gorilla speciation. States HC2, HG, and CG describe the three possible topologies when the human/chimpanzee coalescence occurs prior to the gorilla speciation. Notice that states HC1 and HC2 both have the species topology, but with shallow and deep human/chimpanzee coalescences, respectively, while states HG and CG represent cases of incomplete lineage sorting (ILS). The orangutan is not shown here, but it is used as an outgroup in the analysis. The branch between the human/chimpanzee/gorilla and orangutan ancestors is assumed to be sufficiently long that ILS in this part of the phylogeny can be ignored. (B) The state-transition diagram for the HMM and an example alignment, with alignment blocks colored to match the states that generated them. The transition parameters s, u, and v, which are estimated from the data, reflect the rates of recombinations that convert one genealogy type to another. (Reprinted from Hobolth et al. 2007.)











