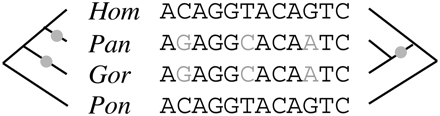
Aligned human (Hum), chimpanzee (Pan), gorilla (Gor), and orangutan (Pon) sequences, showing substitutions (gray) that would each require at least two events to explain under the species phylogeny (left) but only one under a local genealogy resulting from incomplete lineage sorting (right). In a phylogenomic analysis that assumes the species phylogeny holds across the genomes, the observed substitutions will be overcounted, resulting in inflated substitution rates on the branches leading to chimpanzee and gorilla. This type of overcounting can produce complex biases in the prediction of genes or other functional elements, the detection of negative or positive selection, the reconstruction of ancestral genomes, or other phylogenomic analyses (see, e.g., Anisimova et al. 2003).











