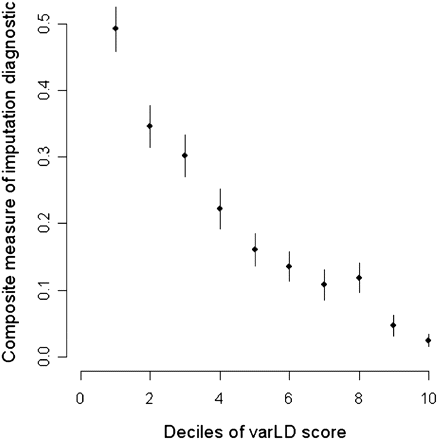
Imputation diagnostics and standardized varLD scores. Comparison of the standardized varLD score against imputation diagnostics generated by IMPUTE when the HapMap YRI is used as a reference panel against Gambian Jola data. The imputation algorithm calculates a measure of information and a confidence score based on the average maximum posterior probability, which we used as surrogates of imputation accuracy. A composite measure of imputation accuracy as measured by the product of call rate and genotype concordance is calculated for the 10 deciles of varLD scores found in the top 20th percentile of the genome-wide distribution of varLD scores. As concordance is measured as the proportion of agreement between the imputed and observed genotypes for the Gambian Jola samples, we only consider autosomal SNPs on the Affymetrix array that are found in the regions identified by varLD.











